Beta Version 0.3.6
We would like to thank all those who have given feedback on Apollo 3 since our
initial beta release of version v0.3.0 in December. We have continued to work
on improving the experience for Apollo 3 users over the course of several
versions, and we'd like to highlight some of the new features we've completed
between then and our newest current release, version v0.3.6.
New features
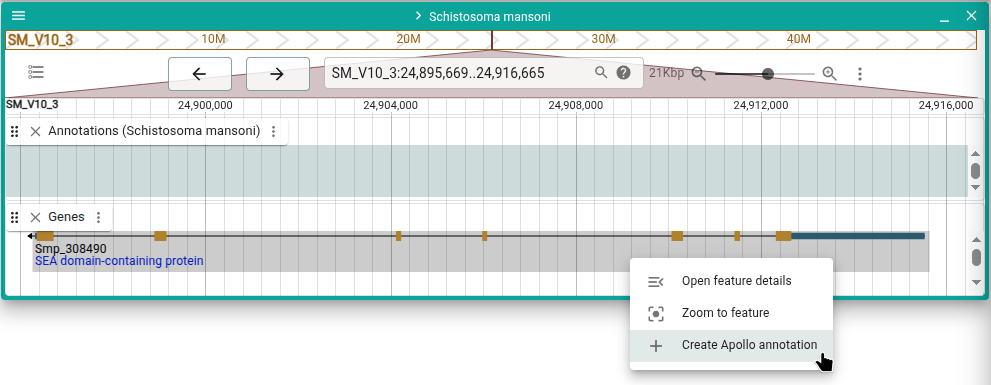
Add feature from JBrowse 2 gene track
Right-click on a feature such as a gene in a JBrowse 2 evidence track and select "Create Apollo annotation" to create a new Apollo annotation from that feature. Attributes on the feature are also copied into the new Apollo annotation.
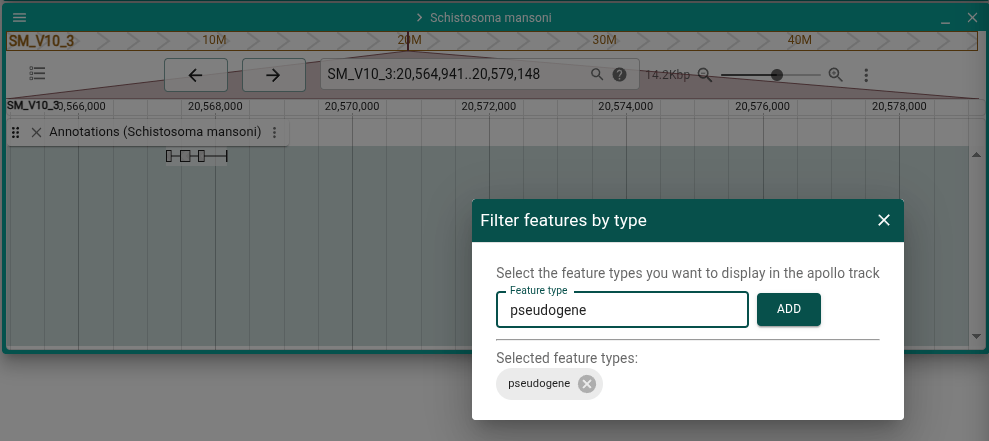
Filter visible features in the display
From the track menu, select "Filter features by type" to filter what features are available in the Apollo track. This could be used, for example, when you want to focus on a subset of all your annotations, such as pseudogenes.
Export GFF3 using Apollo CLI
Previously GFF3 export was only available from the Apollo menu in the UI. Now it can also be done with the Apollo CLI.
apollo export gff3 myAssembly > out.gff3
Protein sequence now included in transcript details
The "Sequence" section of a transcript's details widget now includes the option to view the protein sequence.
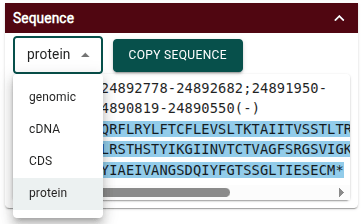
For any of the sequence viewing options, the sequence is in FASTA format. When you use the "Copy sequence" button, the FASTA sequence can be pasted in plain text format (e.g. in a text editor) or with the color highlights (e.g. in Google Docs).
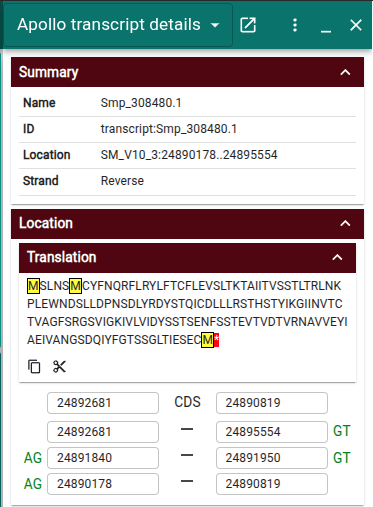
Improved transcript details widget
The transcript details widget now includes a more compact summary of the transcript. It also includes the transcript's protein translation with start and stop codons highlighted as well as the ability to "trim" the CDS to match the nearest start and stop codons if needed with a single click.
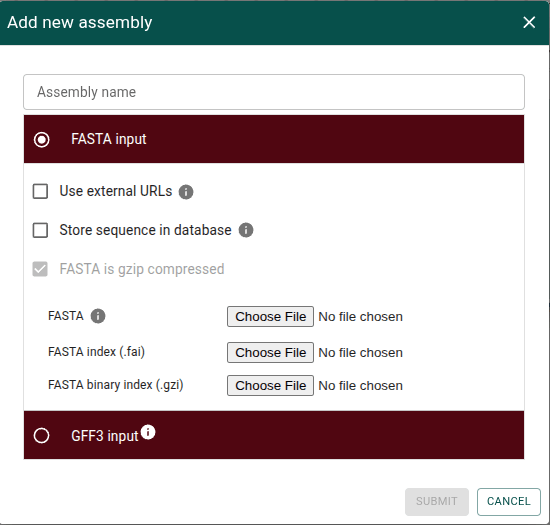
More options available in "Add new assembly" in the UI
The "Add new assembly" dialog in the UI has been improved to add functionality that was previously only available in the Apollo CLI.
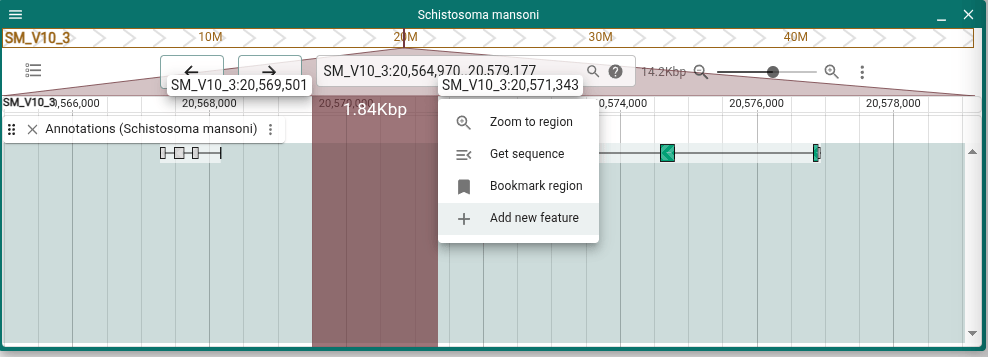
Shortcuts in "Add a feature"
When adding a new feature from scratch, there is now a shortcut to add a gene or transcript with all the necessary subfeatures in a single action.
Access the "Add a feature" dialog by selecting an area in the view ruler track and choosing "Add new feature" from the menu.
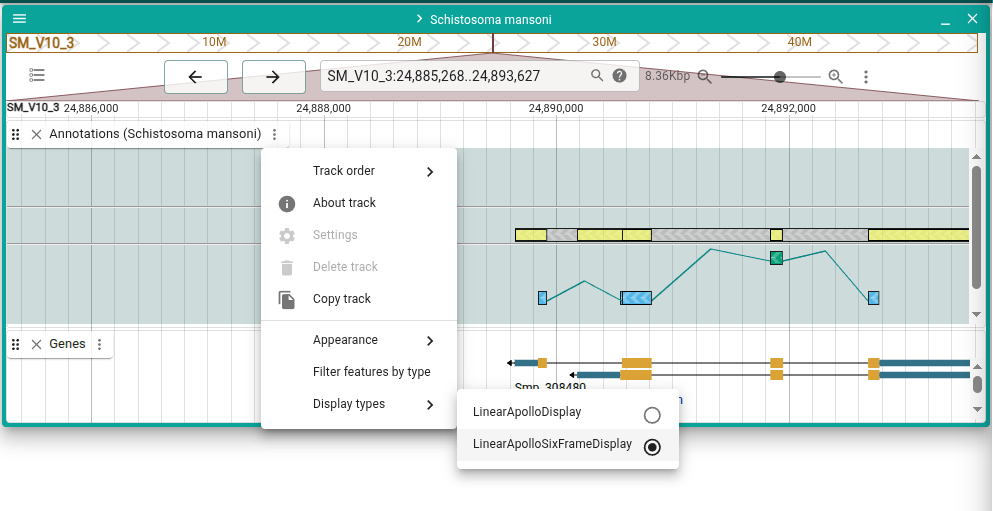
Six frame display
A new display type, the Apollo Six-frame Display, is available for Apollo tracks. It can be accessed from the track menu under "Display types."
This type of view is inspired by the Artemis genome annotation tool and will continue to be updated with more functionality.
Improved functionality
Sequence ontology used to compare equivalent terms
Improvements have been made in using the the configured feature ontology (by default the Sequence Ontology) to detect valid terms for various annotation structures. Any valid ontology terms can be used. For example, you could use the term "exon_of_single_exon_gene" instead of "exon" and Apollo will automatically recognize that as a valid term for the child of a transcript without any additional configuration.
Non-coding genes now render with a gene glyph
Genes without a coding sequence, such as pseudogenes or ncRNA genes, now have improved rendering to match that of coding genes.
Upcoming
Customization
We are working on expanding the options to customize Apollo to fit your own needs. Stay tuned for more information on this soon.
